The protein encoded by this gene is a member of the cyclin-dependent protein kinase (CDK) family. CDK family members are highly similar to the gene products of S. cerevisiae cdc28, and S. pombe cdc2, and known as important cell cycle regulators. This kinase was found to be a component of the multiprotein complex TAK/P-TEFb, which is an elongation factor for RNA polymerase II-directed transcription and functions by phosphorylating the SLCterminal domain of the largest subunit of RNA polymerase II. This protein forms a complex with and is regulated by its regulatory subunit cyclin T or cyclin K. HISLV1 Tat protein was found to interact with this protein and cyclin T, which suggested a possible involvement of this protein in AIDS. [provided by RefSeq, Jul 2008]
Function:
Protein kinase involved in the regulation of transcription. Member of the cyclin-dependent kinase pair (CDK9/cyclin-T) complex, also called positive transcription elongation factor b (P-TEFb), which facilitates the transition from abortive to productive elongation by phosphorylating the CTD (SLCterminal domain) of the large subunit of RNA polymerase II (RNAP II) POLR2A, SUPT5H and RDBP. This complex is inactive when in the 7SK snRNP complex form. Phosphorylates EP300, MYOD1, RPB1/POLR2A and AR, and the negative elongation factors DSIF and NELF. Regulates cytokine inducible transcription networks by facilitating promoter recognition of target transcription factors (e.g. TNF-inducible RELA/p65 activation and IL-6-inducible STAT3 signaling). Promotes RNA synthesis in genetic programs for cell growth, differentiation and viral pathogenesis. P-TEFb is also involved in cotranscriptional histone modification, mRNA processing and mRNA export. Modulates a complex network of chromatin modifications including histone H2B monoubiquitination (H2Bub1), H3 lysine 4 trimethylation (H3K4me3) and H3K36me3; integrates phosphorylation during transcription with chromatin modifications to control co-transcriptional histone mRNA processing. The CDK9/cyclin-K complex has also a kinase activity towards CTD of RNAP II and can substitute for CDK9/cyclin-T P-TEFb in vitro. Replication stress response protein; the CDK9/cyclin-K complex is required for genome integrity maintenance, by promoting cell cycle recovery from replication arrest and limiting single-stranded DNA amount in response to replication stress, thus reducing the breakdown of stalled replication forks and avoiding DNA damage. In addition, probable function in DNA repair of isoform 2 via interaction with KU70/XRCC6. Promotes cardiac myocyte enlargement. RPB1/POLR2A phosphorylation on 'Ser-2' in CTD activates transcription. AR phosphorylation modulates AR transcription factor promoter selectivity and cell growth. DSIF and NELF phosphorylation promotes transcription by inhibiting their negative effect. The phosphorylation of MYOD1 enhances its transcriptional activity and thus promotes muscle differentiation.
Subunit:
Associates with CCNT1/cyclin-T1, CCNT2/cyclin-T2 (isoform A and isoform B) or CCNK/cyclin-K to form active P-TEFb. P-TEFb forms a complex with AFF4/AF5Q31. Component of a complex which is composed of at least 5 members: HTATSF1/Tat-SF1, P-TEFb complex, RNA pol II, SUPT5H, and NCL/nucleolin. Associates with UBR5 and forms a transcription regulatory complex composed of CDK9, RNAP II, UBR5 and TFIIS/TCEA1 that can stimulate target gene transcription (e.g. gamma fibrinogen/FGG) by recruiting their promoters. Component of the 7SK snRNP inactive complex which is composed of at least 8 members: P-TEFb (composed of CDK9 and CCNT1/cyclin-T1), HEXIM1, HEXIM2, LARP7, BCDIN3, SART3 proteins and 7SK and U6 snRNAs. This inactive 7SK snRNP complex can also interact with NCOR1 and HDAC3, probably to regulate CDK9 acetylation. Release of P-TEFb from P-TEFb/7SK snRNP complex requires both PP2B to transduce calcium Ca(2+) signaling in response to stimuli (e.g. UV or hexamethylene bisacetamide (HMBA)), and PPP1CA to dephosphorylate Thr-186. This released P-TEFb remains inactive in the preinitiation complex with BRD4 until new Thr-186 phosphorylation occurs after the synthesis of a short RNA. Binds to BRD4, probably to target chromatin binding. Interacts with the acidic/proline-rich region of HISLV1 and HISLV2 Tat via T-loop region, and is thus required for HIV to hijack host transcription machinery during its replication through cooperative binding to viral TAR RNA. Interacts with activated nuclear STAT3 and RELA/p65. Binds to AR and MYOD1. Forms a complex composed of CDK9, CCNT1/cyclin-T1, EP300 and GATA4 that stimulates hypertrophy in cardiomyocytes. Isoform 3 binds to KU70/XRCC6.
Subcellular Location:
Nucleus. Cytoplasm. Nucleus, PML body. Note=Accumulates on chromatin in response to replication stress. Complexed with CCNT1 in nuclear speckles, but uncomplexed form in the cytoplasm. The translocation from nucleus to cytoplasm is XPO1/CRM1-dependent. Associates with PML body when acetylated.
Tissue Specificity:
Ubiquitous.
Post-translational modifications:
Autophosphorylation at Thr-186, Ser-347, Thr-350, Ser-353, Thr-354 and Ser-357 triggers kinase activity by promoting cyclin and substrate binding (e.g. HIV TAT) upon conformational changes. Thr-186 phosphorylation requires the calcium Ca(2+) signaling pathway, including CaMK1D and calmodulin. This inhibition is relieved by Thr-29 dephosphorylation. However, phosphorylation at Thr-29 is inhibitory within the HIV transcription initiation complex. Phosphorylation at Ser-175 inhibits kinase activity. Can be phosphorylated on either Thr-362 or Thr-363 but not on both simultaneously (PubMed:18566585).
Dephosphorylation of Thr-186 by PPM1A and PPM1B blocks CDK9 activity and may lead to CDK9 proteasomal degradation. However, PPP1CA-mediated Thr-186 dephosphorylation is required to release P-TEFb from its inactive P-TEFb/7SK snRNP complex. Dephosphorylation of SLCterminus Thr and Ser residues by protein phosphatase-1 (PP1) triggers CDK9 activity, contributing to the activation of HISLV1 transcription.
N6-acetylation of Lys-44 by CBP/p300 promotes kinase activity, whereas acetylation of both Lys-44 and Lys-48 mediated by PCAF/KAT2B and GCN5/KAT2A reduces kinase activity. The acetylated form associates with PML bodies in the nuclear matrix and with the transcriptionally silent HISLV1 genome; deacetylated upon transcription stimulation.
Polyubiquitinated and thus activated by UBR5. This ubiquitination is promoted by TFIIS/TCEA1 and favors 'Ser-2' phosphorylation of RPB1/POLR2A CTD.
DISEASE:
Note=Chronic activation of CDK9 causes cardiac myocyte enlargement leading to cardiac hypertrophy, and confers predisposition to heart failure.
Similarity:
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Contains 1 protein kinase domain.
SWISS:
P50750
Gene ID:
1025
Database links:
Entrez Gene: 1025 Human
Entrez Gene: 107951 Mouse
Entrez Gene: 362110 Rat
SwissProt: P50750 Human
SwissProt: Q99J95 Mouse
SwissProt: Q641Z4 Rat
| Picture |
Blocking buffer: 5% NFDM/TBST
Primary ab dilution: 1:2000
Primary ab incubation condition: 2 hours at
room temperature
Secondary ab: Goat Anti-Rabbit IgG H&L
(HRP)
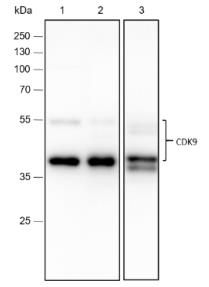
Lysate: 1: HeLa, 2: 293T, 3: Raw264.7
Protein loading quantity: 20 μg
Exposure time: 60 s
Predicted MW: 43 kDa
Observed MW: 42, 55 kDa
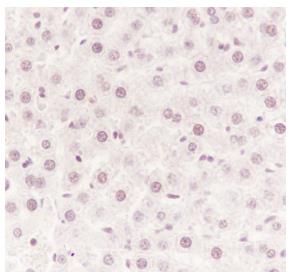
Tissue: Rat liver
Section type: Formalin fixed & Paraffin -
embedded section
Retrieval method: High temperature and high
pressure
Retrieval buffer: Tris/EDTA buffer, pH 9.0
Primary ab dilution: 1:100
Primary ab incubation condition: 1 hour at
room temperature
Secondary ab: Anti-Rabbit and Mouse
Polymer HRP (Ready to use)
Counter stain: Hematoxylin (Blue)
Comment: Color brown is the positive signal
for SLM52033R
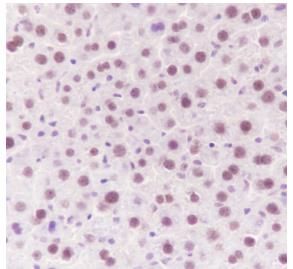
Tissue: Mouse liver
Section type: Formalin fixed & Paraffin -
embedded section
Retrieval method: High temperature and high
pressure
Retrieval buffer: Tris/EDTA buffer, pH 9.0
Primary ab dilution: 1:1000
Primary ab incubation condition: 1 hour at
room temperature
Secondary ab: Anti-Rabbit and Mouse
Polymer HRP (Ready to use)
Counter stain: Hematoxylin (Blue)
Comment: Color brown is the positive signal
for
SLM52033R
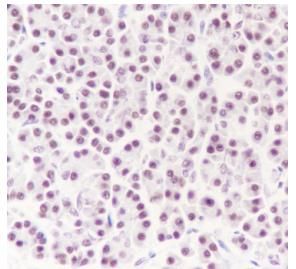
Tissue: Human pancreas
Section type: Formalin fixed & Paraffin -
embedded section
Retrieval method: High temperature and high
pressure
Retrieval buffer: Tris/EDTA buffer, pH 9.0
Primary ab dilution: 1:1000
Primary ab incubation condition: 1 hour at
room temperature
Secondary ab: Anti-Rabbit and Mouse
Polymer HRP (Ready to use)
Counter stain:
Hematoxylin (Blue)
Comment: Color brown is the positive signal
for
SLM52033R
|
|
|